Global DNA hypomethylation in canine mammary tumors
Abstract
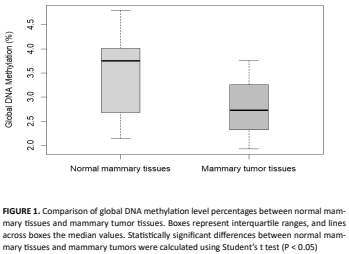
Due to its influence in transcriptional potential of genes, genetic regulation by means of epigenetic mechanisms is essential for normal growth and development. In mammary cancer, epigenetic modifications play a key role for its development and progression. In early carcinogenesis stages, due to genetic alterations or environmental factors, chromatin structure alterations due to DNA methylation and post-translational modifications of DNA-bound proteins may appear. As with other types of tumor, genome-wide hypomethylation and hypermethylation of specific genes, particularly in CpG islands that normally are not methylated, are observed. In order to compare global DNA methylation levels between tumor tissue and normal mammary tissue, we studied 11 intact female dogs with mammary tumors. Both types of tissue were collected during surgery, with subsequent clinical staging and histopathological classification of tumors. For each animal, DNA was extracted from paired samples of tumor tissue and normal mammary tissue. Global genomic methylation levels were calculated by relative quantitation of 5-methyl-2’-deoxycytidine (5mdC) with HPLC. Results showed that tumoral tissue had a global DNA hypomethylation when compared with normal mammary tissue (P < 0.05). This difference was greater in high histopathological grade tumors, characterized by their aggressive clinical behavior and high metastatic rate. These findings underscore the importance of additional studies in this line of research, with greater sample sizes. In the future, global DNA methylation may be used as a prognosis biomarker for mammary cancer in dogs.
Downloads
References
Zhou WM, Liu B, Shavandi A, Li L, Song H, Zhang JY. Methylation landscape: Targeting writer or eraser to discover anti-cancer drug. Front Pharmacol. [Internet]. 2021; 12:690057. doi: https://doi.org/qf7c DOI: https://doi.org/10.3389/fphar.2021.690057
Karsli-Ceppioglu S, Dagdemir A, Judes G, Ngollo M, Penault-Llorca F, Pajon A, Bignon YJ, Bernard-Gallon D. Epigenetic mechanisms of breast cancer: an update of the current knowledge. Epigenomics. [Internet]. 2014; 6(6):651-664. doi: https://doi.org/gms37n DOI: https://doi.org/10.2217/epi.14.59
Bártová E. Epigenetic and gene therapy in human and veterinary medicine. Environ. Epigenetics. [Internet]. 2024; 10(1):dvae006. doi: https://doi.org/qf7f DOI: https://doi.org/10.1093/eep/dvae006
Szczepanek J, Skorupa M, Jarkiewicz-Tretyn J, Cybulski C, Tretyn A. Harnessing Epigenetics for Breast Cancer Therapy: The Role of DNA Methylation, Histone Modifications, and MicroRNA. Int. J. Mol. Sci. [Internet]. 2023; 24(8):7235. doi: https://doi.org/qf7h DOI: https://doi.org/10.3390/ijms24087235
Brookes E, Shi Y. Diverse epigenetic mechanisms of human disease. Annu. Rev. Genet. [Internet]. 2014; 48:237-268. doi: https://doi.org/qf7j DOI: https://doi.org/10.1146/annurev-genet-120213-092518
Nishiyama A, Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends Genet. [Internet]. 2021; 37(11):1012-1027. doi: https://doi.org/gm5thv DOI: https://doi.org/10.1016/j.tig.2021.05.002
Menezo Y, Silvestris E, Dale B, Elder K. Oxidative stress and alterations in DNA methylation: two sides of the same coin in reproduction. Reprod. Biomed. Online. [Internet]. 2016; 33(6):668-683. doi: https://doi.org/f9p3tc DOI: https://doi.org/10.1016/j.rbmo.2016.09.006
Pasculli B, Barbano R, Parrella P. Epigenetics of breast cancer: Biology and clinical implication in the era of precision medicine. Semin. Cancer Biol. [Internet]. 2018; 51:22-35. doi: https://doi.org/gd45tv DOI: https://doi.org/10.1016/j.semcancer.2018.01.007
Wu Y, Sarkissyan M, Vadgama JV. Epigenetics in Breast and Prostate Cancer. In: Verma, M. (eds). Cancer Epigenetics. Methods Mol. Biol. [Internet]. 2015; 1238:425–466. doi: https://doi.org/f6trhx DOI: https://doi.org/10.1007/978-1-4939-1804-1_23
Győrffy B, Bottai G, Fleischer T, Munkácsy G, Budczies J, Paladini L, Børresen-Dale A, Kristensen VN, Santarpia L. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int. J. Cancer. [Internet]. 2016; 138(1):87-97. doi: https://doi.org/gjjd2s DOI: https://doi.org/10.1002/ijc.29684
Berdasco M, Fraga M, Esteller M. Quantification of global DNA methylation by capillary electrophoresis and mass spectrometry. In: Tost, J. (eds) DNA Methylation. Methods Mol. Biol. [Internet]. 2009; 507:23–34. doi: https://doi.org/cjz5p9 DOI: https://doi.org/10.1007/978-1-59745-522-0_2
Cappetta M, Berdasco M, Hochmann J, Bonilla C, Sans M,Hidalgo PC, Artagaveytia N, Kittles R, Martínez M, Esteller M, Bertoni B. Effect of genetic ancestry on leukocyte global DNA methylation in cancer patients. BMC Cancer. [Internet]. 2015; 15:434. doi: https://doi.org/f7dp9f DOI: https://doi.org/10.1186/s12885-015-1461-0
Xavier P, Müller S, Fukumasu H. Epigenetic Mechanisms in Canine Cancer. Front. Oncol. [Internet]. 2020; 10:591843. doi: https://doi.org/qf7k DOI: https://doi.org/10.3389/fonc.2020.591843
Abdelmegeed S, Mohammed S. Canine mammary tumors as a model for human disease (Review). Oncol. Lett. [Internet] 2018; 15(6):8195–8205. doi: https://doi.org/mdsg DOI: https://doi.org/10.3892/ol.2018.8411
Jeong S, Lee K, Nam A, Cho J. Genome-wide methylation profiling in canine mammary tumor reveals miRNA candidates associated with human breast cancer. Cancers. [Internet] 2019; 11(10):1466. doi: https://doi.org/qf7m DOI: https://doi.org/10.3390/cancers11101466
Biondi L, Tedardi M, Gentile L, Chamas P, Dagli M. Quantification of global DNA methylation in canine mammary gland tumors via immunostaining of 5-methylcytosine: Histopathological and clinical correlations. Front. Vet. Sci. [Internet] 2021; 8:628241. doi: https://doi.org/qf7n DOI: https://doi.org/10.3389/fvets.2021.628241
Brandão Y, Toledo M, Chequin A, Cristo T, Sousa R, Ramos E, Klassen G. DNA methylation status of the estrogen receptor α gene in canine mammary tumors. Vet Pathol. [Internet] 2018; 55(4):510–516. doi: https://doi.org/qf7p DOI: https://doi.org/10.1177/0300985818763711
Beetch M, Harandi S, Yang T, Boycott C, Chen Y, Stefanskan B, Mohammed S. DNA methylation landscape of triple- negative ductal carcinoma in situ (DCIS) progressing to the invasive stage in canine breast cancer. Sci. Rep. [Internet] 2020; 10:2415. https://doi.org/qf7q DOI: https://doi.org/10.1038/s41598-020-59260-4
Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. New York University: Lawrence Erlbaum Associates. 1988.
Goldshmidt M, Peña L, Rasotto R, Zappulli, V. Classification and grading of canine mammary tumors. Vet. Pathol. [Internet] 2011; 48(1):117–131. doi: https://doi.org/cg36bk DOI: https://doi.org/10.1177/0300985810393258
Egenvall A, Bonnett B, Ohagen P, Olson P, Hedhammar A, von Euler H. Incidence of and survival after mammary tumors in a population of over 80,000 insured female dogs in Sweden from 1995 to 2002. Prevent. Vet. Med. [Internet]. 2005; 69(1-2):109–127. doi: https://doi.org/fw7k7z DOI: https://doi.org/10.1016/j.prevetmed.2005.01.014
Hillyar C, Rallis K, Varghese J. Advances in epigenetic cancer therapeutics. Cureus. [Internet] 2020; 12(11):e11725. doi: https://doi.org/gqsrqz DOI: https://doi.org/10.7759/cureus.11725